pub struct Block<const TRACE: bool, const X_DROP: bool = false, const LOCAL_START: bool = false, const FREE_QUERY_START_GAPS: bool = false, const FREE_QUERY_END_GAPS: bool = false> { /* private fields */ }Expand description
Data structure storing the settings for Block Aligner.
A diagram showing different ways Block Aligner can be used:
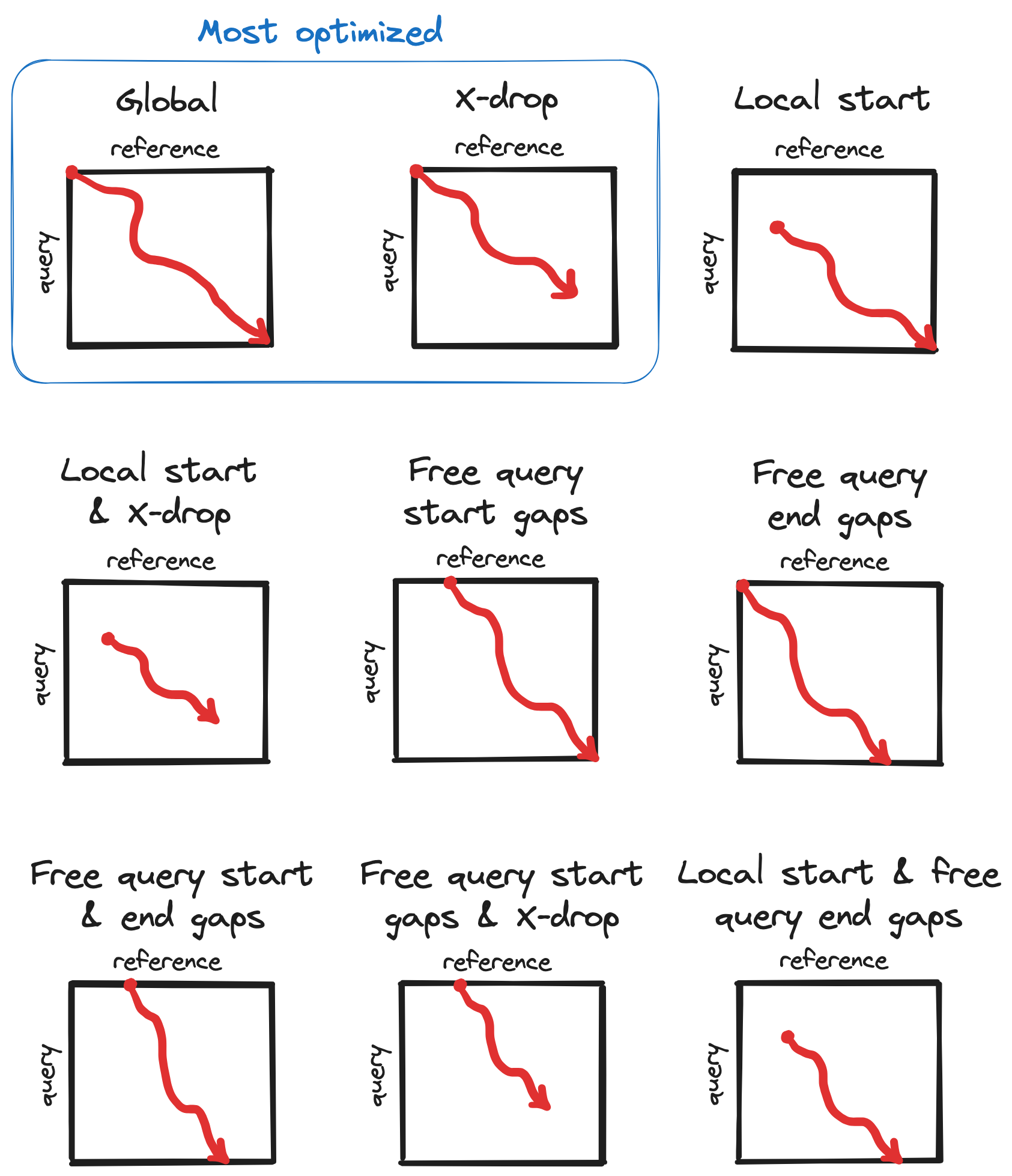
Implementations§
Source§impl<const TRACE: bool, const X_DROP: bool, const LOCAL_START: bool, const FREE_QUERY_START_GAPS: bool, const FREE_QUERY_END_GAPS: bool> Block<TRACE, X_DROP, LOCAL_START, FREE_QUERY_START_GAPS, FREE_QUERY_END_GAPS>
impl<const TRACE: bool, const X_DROP: bool, const LOCAL_START: bool, const FREE_QUERY_START_GAPS: bool, const FREE_QUERY_END_GAPS: bool> Block<TRACE, X_DROP, LOCAL_START, FREE_QUERY_START_GAPS, FREE_QUERY_END_GAPS>
Sourcepub fn new(query_len: usize, reference_len: usize, max_size: usize) -> Self
pub fn new(query_len: usize, reference_len: usize, max_size: usize) -> Self
Allocate a block aligner instance with an upper bound query length, reference length, and max block size.
A block aligner instance can be reused for multiple alignments as long as the aligned sequence lengths and block sizes do not exceed the specified upper bounds.
Sourcepub fn align<M: Matrix>(
&mut self,
query: &PaddedBytes,
reference: &PaddedBytes,
matrix: &M,
gaps: Gaps,
size: RangeInclusive<usize>,
x_drop: i32,
)
pub fn align<M: Matrix>( &mut self, query: &PaddedBytes, reference: &PaddedBytes, matrix: &M, gaps: Gaps, size: RangeInclusive<usize>, x_drop: i32, )
Align two sequences with block aligner.
If TRACE is true, then information for computing the traceback will be stored.
After alignment, the traceback CIGAR string can then be computed.
This will slow down alignment and use a lot more memory.
If X_DROP is true, then the alignment process will be terminated early when
the max score in the current block drops by x_drop below the max score encountered
so far. The location of the max score is stored in the alignment result.
This allows the alignment to end anywhere in the DP matrix.
If X_DROP is false, then global alignment is done.
If LOCAL_START is true, then the alignment is allowed to start anywhere in the DP matrix.
Local alignment can be accomplished by setting LOCAL_START and X_DROP to true and x_drop
to a very large value.
If FREE_QUERY_START_GAPS is true, then gaps before the start of the query are free.
If FREE_QUERY_END_GAPS is true, then gaps after the end of the query are free.
Note that this has a limitation: the min block size must be greater than the length of the query.
Since larger scores are better, gap and mismatches penalties must be negative.
The minimum and maximum sizes of the block must be powers of 2 that are greater than the number of 16-bit lanes in a SIMD vector.
The block aligner algorithm will dynamically shift a block down or right and grow its size to efficiently calculate the alignment between two strings. This is fast, but it may be slightly less accurate than computing the entire the alignment dynamic programming matrix. Growing the size of the block allows larger gaps and other potentially difficult regions to be handled correctly. The algorithm also allows shrinking the block size for greater efficiency when handling regions in the sequences with no gaps. 16-bit deltas and 32-bit offsets are used to ensure that accurate scores are computed, even when the the strings are long.
When aligning sequences q against r, this algorithm computes cells in the DP matrix
with |q| + 1 rows and |r| + 1 columns.
X-drop alignment with ByteMatrix is not supported.
Sourcepub fn align_exp<M: Matrix>(
&mut self,
query: &PaddedBytes,
reference: &PaddedBytes,
matrix: &M,
gaps: Gaps,
size: RangeInclusive<usize>,
x_drop: i32,
target_score: i32,
) -> Option<usize>
pub fn align_exp<M: Matrix>( &mut self, query: &PaddedBytes, reference: &PaddedBytes, matrix: &M, gaps: Gaps, size: RangeInclusive<usize>, x_drop: i32, target_score: i32, ) -> Option<usize>
Align two sequences with exponential search on the min block size.
This calls align multiple times, doubling the min block size in each iteration
until either the max block size is reached or the score reaches or exceeds the target score.
Sourcepub fn align_profile<P: Profile>(
&mut self,
query: &PaddedBytes,
profile: &P,
size: RangeInclusive<usize>,
x_drop: i32,
)
pub fn align_profile<P: Profile>( &mut self, query: &PaddedBytes, profile: &P, size: RangeInclusive<usize>, x_drop: i32, )
Align a sequence to a profile with block aligner.
If TRACE is true, then information for computing the traceback will be stored.
After alignment, the traceback CIGAR string can then be computed.
This will slow down alignment and use a lot more memory.
If X_DROP is true, then the alignment process will be terminated early when
the max score in the current block drops by x_drop below the max score encountered
so far. The location of the max score is stored in the alignment result.
This allows the alignment to end anywhere in the DP matrix.
If X_DROP is false, then global alignment is done.
If LOCAL_START is true, then the alignment is allowed to start anywhere in the DP matrix.
Local alignment can be accomplished by setting LOCAL_START and X_DROP to true and x_drop
to a very large value.
If FREE_QUERY_START_GAPS is true, then gaps before the start of the query are free.
If FREE_QUERY_END_GAPS is true, then gaps after the end of the query are free.
Note that this has a limitation: the min block size must be greater than the length of the query.
Since larger scores are better, gap and mismatches penalties must be negative.
The minimum and maximum sizes of the block must be powers of 2 that are greater than the number of 16-bit lanes in a SIMD vector.
The block aligner algorithm will dynamically shift a block down or right and grow its size to efficiently calculate the alignment between two strings. This is fast, but it may be slightly less accurate than computing the entire the alignment dynamic programming matrix. Growing the size of the block allows larger gaps and other potentially difficult regions to be handled correctly. The algorithm also allows shrinking the block size for greater efficiency when handling regions in the sequences with no gaps. 16-bit deltas and 32-bit offsets are used to ensure that accurate scores are computed, even when the the strings are long.
When aligning sequence q against profile p, this algorithm computes cells in the DP matrix
with |q| + 1 rows and |p| + 1 columns.
Sourcepub fn align_profile_exp<P: Profile>(
&mut self,
query: &PaddedBytes,
profile: &P,
size: RangeInclusive<usize>,
x_drop: i32,
target_score: i32,
) -> Option<usize>
pub fn align_profile_exp<P: Profile>( &mut self, query: &PaddedBytes, profile: &P, size: RangeInclusive<usize>, x_drop: i32, target_score: i32, ) -> Option<usize>
Align a sequence to a profile with exponential search on the min block size.
This calls align_profile multiple times, doubling the min block size in each iteration
until either the max block size is reached or the score reaches or exceeds the target score.
Sourcepub fn res(&self) -> AlignResult
pub fn res(&self) -> AlignResult
Get the resulting score and ending location of the alignment.