Peroxide
Pure Rust numeric library contains linear algebra, numerical analysis, statistics and machine learning tools with R, MATLAB, Python like macros.
Latest README version
Corresponding to 0.12.3.
Pre-requisite
- Python module :
matplotlibfor plotting
Install
- Add next line to your
cargo.toml
= "0.12"
Module Structure
- src
- lib.rs :
modandre-export - macros : Macro files
- julia_macro.rs : Julia like macro
- matlab_macro.rs : MATLAB like macro
- mod.rs
- r_macro.rs : R like macro
- ml : For machine learning (Beta)
- numerical : To do numerical things
- bdf.rs : Backward Differentiation Formula
- gauss_legendre.rs : Gauss-Legendre 4th order
- interp.rs : Interpolation
- mod.rs
- newton.rs : Newton's Method
- ode.rs : Main ODE solver
- optimize.rs : Non-linear regression
- spline.rs : Natural Spline
- utils.rs : Utils to do numerical things (e.g. jacobian)
- operation : To define general operations
- extra_ops.rs : Missing operations & Real Trait
- mut_ops.rs : Mutable operations
- mod.rs
- number.rs : Number type (include
f64,Dual)
- special : Wrapper for
specialcrate- mod.rs
- function.rs : Special functions
- statistics : Statistical Tools
- structure : Fundamental data structures
- dataframe.rs : Not yet implemented
- dual.rs : Dual number system for automatic differentiation
- hyper_dual.rs : Hyper dual number system for automatic differentiation
- matrix.rs : Matrix
- multinomial.rs : For multinomial (Beta)
- mod.rs
- polynomial.rs : Polynomial
- vector.rs : Extra tools for
Vec<f64>
- util
- mod.rs
- api.rs : Matrix constructor for various language style
- non_macro.rs : Primordial version of macros
- pickle.rs : To handle
pickledata structure - plot.rs : To draw plot (using
pyo3) - print.rs : To print conveniently
- useful.rs : Useful utils to implement library
- writer.rs : More convenient write system
- lib.rs :
Documentation
Covered Contents
- Linear Algebra
- Effective Matrix structure
- Transpose, Determinant, Diagonal
- LU Decomposition, Inverse matrix, Block partitioning
- Column, Row operations
- Functional Programming
- More easy functional programming with
Vec<f64> - For matrix, there are three maps
fmap: map for all elementscol_map: map for column vectorsrow_map: map for row vectors
- More easy functional programming with
- Automatic Differentiation
- Dual number system - for 1st order AD
- Hyper dual number system - for 2nd order AD
- Exact jacobian
Realtrait to constrain forf64andDualNumberstructure to unifyf64andDual
- Numerical Analysis
- Lagrange interpolation
- Cubic spline
- Non-linear regression
- Gradient Descent
- Gauss Newton
- Levenberg Marquardt
- Ordinary Differential Equation
- Euler
- Runge Kutta 4th order
- Backward Euler
- Gauss Legendre 4th order
- Statistics
- More easy random with
randcrate - Probability Distributions
- Bernoulli
- Uniform
- Normal
- Gamma
- Beta
- RNG algorithms
- Acceptance Rejection
- Marsaglia Polar
- Ziggurat
- More easy random with
- Special functions
- Wrapper for
specialcrate
- Wrapper for
- Utils
- R-like macro & functions
- Matlab-like macro & functions
- Numpy-like macro & functions
- Julia-like macro & functions
- Plotting
- With
pyo3&matplotlib
- With
Example
Basic Runge-Kutta 4th order with inline-python
extern crate peroxide;
extern crate inline_python;
use *;
use python;
// dy/dx = (5x^2 - y) / e^(x+y)
Basic Runge-Kutta 4th order with advanced plotting
extern crate peroxide;
use *;
Basic Runge-Kutta 4th order with Stop condition
extern crate peroxide;
use *;

Multi-Layer Perceptron (from scratch)
extern crate peroxide;
use *;
// x : n x L
// xb: n x (L+1)
// v : (L+1) x M
// a : n x M
// ab: n x (M+1)
// w : (M+1) x n
// wb: M x N
// y : n x N
// t : n x N
// dh: n x M
// do: n x N
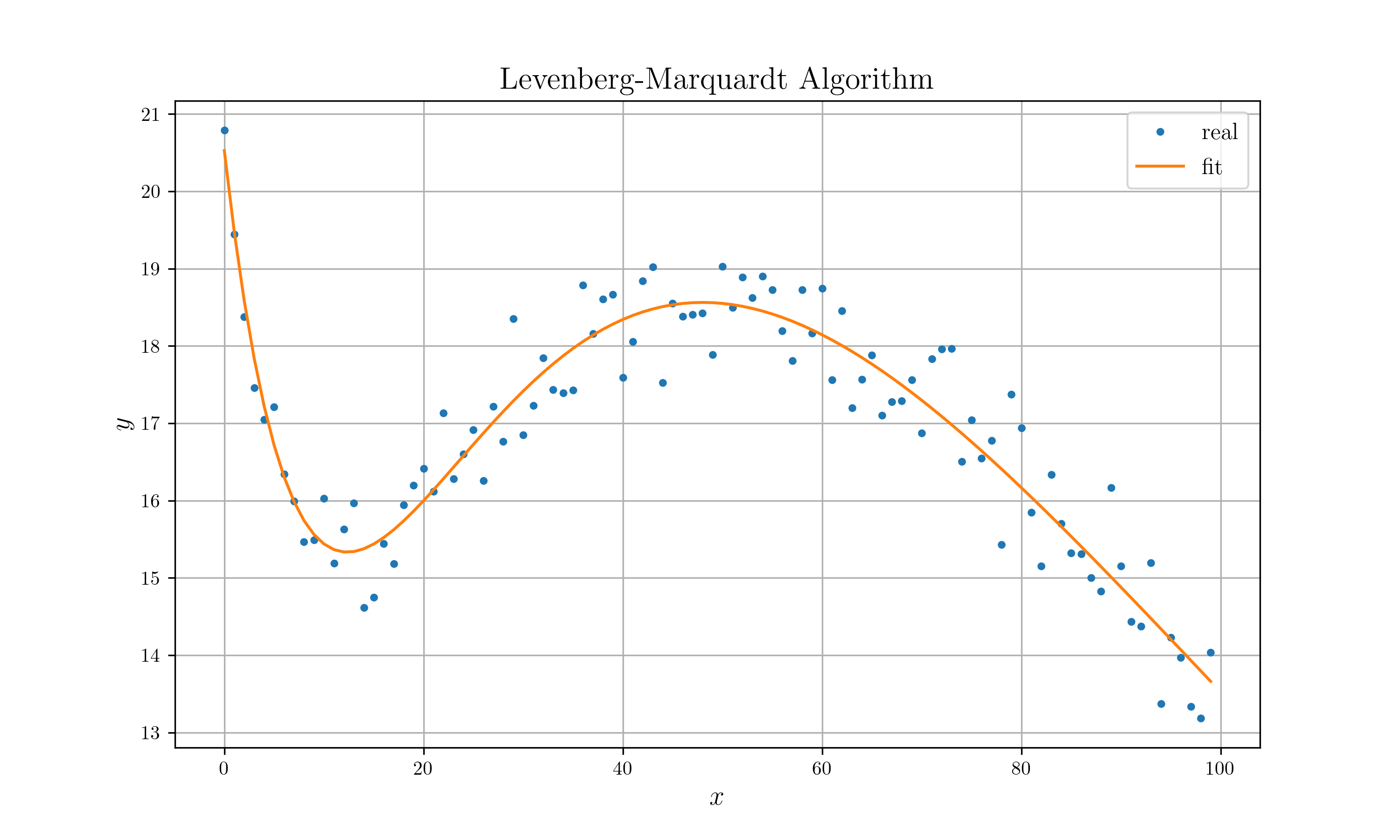
Levenberg-Marquardt Algorithm (refer to lm.pdf)
extern crate peroxide;
use *;
Version Info
To see RELEASES.md
TODO
To see TODO.md

