rebop/lib.rs
1//! rebop is a fast stochastic simulator for well-mixed chemical
2//! reaction networks.
3//!
4//! Performance and ergonomics are taken very seriously. For this reason,
5//! two independent APIs are provided to describe and simulate reaction
6//! networks:
7//!
8//! * a macro-based DSL implemented by [`define_system`], usually the
9//! most efficient, but that requires to compile a rust program;
10//! * a function-based API implemented by the module [`gillespie`], also
11//! available through Python bindings. This one does not require a rust
12//! compilation and allows the system to be defined at run time. It is
13//! typically 2 or 3 times slower than the macro DSL, but still faster
14//! than all other software tried.
15//!
16//! # The macro DSL
17//!
18//! It currently only supports reaction rates defined by the law of mass
19//! action. The following macro defines a dimerization reaction network
20//! naturally:
21//!
22//! ```rust
23//! use rebop::define_system;
24//! define_system! {
25//! r_tx r_tl r_dim r_decay_mRNA r_decay_prot;
26//! Dimers { gene, mRNA, protein, dimer }
27//! transcription : gene => gene + mRNA @ r_tx
28//! translation : mRNA => mRNA + protein @ r_tl
29//! dimerization : 2 protein => dimer @ r_dim
30//! decay_mRNA : mRNA => @ r_decay_mRNA
31//! decay_protein : protein => @ r_decay_prot
32//! }
33//! ```
34//!
35//! To simulate the system, put this definition in a rust code file and
36//! instantiate the problem, set the parameters, the initial values, and
37//! launch the simulation:
38//!
39//! ```rust
40//! # use rebop::define_system;
41//! # define_system! {
42//! # r_tx r_tl r_dim r_decay_mRNA r_decay_prot;
43//! # Dimers { gene, mRNA, protein, dimer }
44//! # transcription : gene => gene + mRNA @ r_tx
45//! # translation : mRNA => mRNA + protein @ r_tl
46//! # dimerization : 2 protein => dimer @ r_dim
47//! # decay_mRNA : mRNA => @ r_decay_mRNA
48//! # decay_protein : protein => @ r_decay_prot
49//! # }
50//! let mut problem = Dimers::new();
51//! problem.r_tx = 25.0;
52//! problem.r_tl = 1000.0;
53//! problem.r_dim = 0.001;
54//! problem.r_decay_mRNA = 0.1;
55//! problem.r_decay_prot = 1.0;
56//! problem.gene = 1;
57//! problem.advance_until(1.0);
58//! println!("t = {}: dimer = {}", problem.t, problem.dimer);
59//! ```
60//!
61//! Or for the classic SIR example:
62//!
63//! ```rust
64//! use rebop::define_system;
65//!
66//! define_system! {
67//! r_inf r_heal;
68//! SIR { S, I, R }
69//! infection : S + I => 2 I @ r_inf
70//! healing : I => R @ r_heal
71//! }
72//!
73//! fn main() {
74//! let mut problem = SIR::new();
75//! problem.r_inf = 1e-4;
76//! problem.r_heal = 0.01;
77//! problem.S = 999;
78//! problem.I = 1;
79//! println!("time,S,I,R");
80//! for t in 0..250 {
81//! problem.advance_until(t as f64);
82//! println!("{},{},{},{}", problem.t, problem.S, problem.I, problem.R);
83//! }
84//! }
85//! ```
86//!
87//! which can produce an output similar to this one:
88//!
89//! 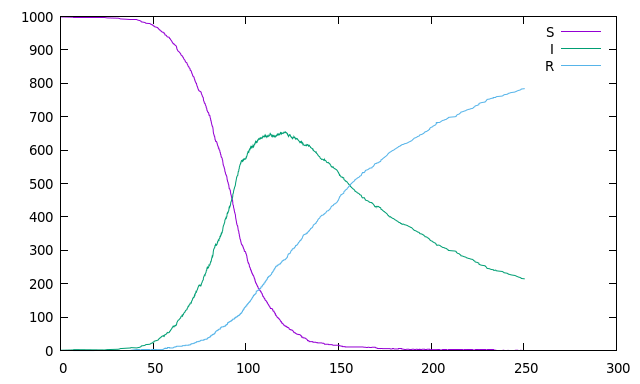
90//!
91//! # Python bindings
92//!
93//! This API shines through the Python bindings which allow one to
94//! define a model easily:
95//!
96//! ```python
97//! import rebop
98//!
99//! sir = rebop.Gillespie()
100//! sir.add_reaction(1e-4, ['S', 'I'], ['I', 'I'])
101//! sir.add_reaction(0.01, ['I'], ['R'])
102//! print(sir)
103//!
104//! ds = sir.run({'S': 999, 'I': 1}, tmax=250, nb_steps=250)
105//! ```
106//!
107//! You can test this code by installing `rebop` from PyPI with
108//! `pip install rebop`. To build the Python bindings from source,
109//! the simplest is to clone this git repository and use `maturin
110//! develop`.
111//!
112//! # The traditional API
113//!
114//! The function-based API underlying the Python package is also available
115//! from Rust, if you want to be able to define models at run time (instead
116//! of at compilation time with the macro DSL demonstrated above).
117//! The SIR model is defined as:
118//!
119//! ```rust
120//! use rebop::gillespie::{Gillespie, Rate};
121//!
122//! let mut sir = Gillespie::new([999, 1, 0], false);
123//! // [ S, I, R]
124//! // S + I => 2 I with rate 1e-4
125//! sir.add_reaction(Rate::lma(1e-4, [1, 1, 0]), [-1, 1, 0]);
126//! // I => R with rate 0.01
127//! sir.add_reaction(Rate::lma(0.01, [0, 1, 0]), [0, -1, 1]);
128//!
129//! println!("time,S,I,R");
130//! for t in 0..250 {
131//! sir.advance_until(t as f64);
132//! println!("{},{},{},{}", sir.get_time(), sir.get_species(0), sir.get_species(1), sir.get_species(2));
133//! }
134//! ```
135//!
136//! # Performance
137//!
138//! Performance is taken very seriously, and as a result, rebop
139//! outperforms every other package and programming language that we
140//! tried.
141//!
142//! *Disclaimer*: Most of this software currently contains much more
143//! features than rebop (e.g. spatial models, custom reaction rates,
144//! etc.). Some of these features might have required them to make
145//! compromises on speed. Moreover, as much as we tried to keep the
146//! comparison fair, some return too much or too little data, or write
147//! them on disk. The baseline that we tried to approach for all these
148//! programs is the following: *the model was just modified, we want
149//! to simulate it `N` times and print regularly spaced measurement
150//! points*. This means that we always include initialization or
151//! (re-)compilation time if applicable. We think that it is the most
152//! typical use-case of a researcher who works on the model. This
153//! benchmark methods allows to record both the initialization time
154//! (y-intercept) and the simulation time per simulation (slope).
155//!
156//! Many small benchmarks on toy examples are tracked to guide the
157//! development. To compare the performance with other software,
158//! we used a real-world model of low-medium size (9 species and 16
159//! reactions): the Vilar oscillator (*Mechanisms of noise-resistance
160//! in genetic oscillators*, Vilar et al., PNAS 2002). Here, we
161//! simulate this model from `t=0` to `t=200`, reporting the state at
162//! time intervals of `1` time unit.
163//!
164//! 
165//!
166//! We can see that rebop's macro DSL is the fastest of all, both in
167//! time per simulation, and with compilation time included. The second
168//! fastest is rebop's traditional API invoked by convenience through
169//! the Python bindings.
170//!
171//! # Features to come
172//!
173//! * compartment volumes
174//! * arbitrary reaction rates
175//! * other SSA algorithms
176//! * tau-leaping
177//! * adaptive tau-leaping
178//! * hybrid models (continuous and discrete)
179//! * SBML
180//! * CLI interface
181//! * parameter estimation
182//! * local sensitivity analysis
183//! * parallelization
184//!
185//! # Features probably not to come
186//!
187//! * events
188//! * space (reaction-diffusion systems)
189//! * rule modelling
190//!
191//! # Benchmark ideas
192//!
193//! * DSMTS
194//! * purely decoupled exponentials
195//! * ring
196//! * Toggle switch
197//! * LacZ, LacY/LacZ (from STOCKS)
198//! * Lotka Volterra, Michaelis--Menten, Network (from StochSim)
199//! * G protein (from SimBiology)
200//! * Brusselator / Oregonator (from Cellware)
201//! * GAL, repressilator (from Dizzy)
202//!
203//! # Similar software
204//!
205//! ## Maintained
206//!
207//! * [GillesPy2](https://github.com/StochSS/GillesPy2)
208//! * [STEPS](https://github.com/CNS-OIST/STEPS)
209//! * [SimBiology](https://fr.mathworks.com/help/simbio/)
210//! * [Copasi](http://copasi.org/)
211//! * [BioNetGen](http://bionetgen.org/)
212//! * [VCell](http://vcell.org/)
213//! * [Smoldyn](http://www.smoldyn.org/)
214//! * [KaSim](https://kappalanguage.org/)
215//! * [StochPy](https://github.com/SystemsBioinformatics/stochpy)
216//! * [BioSimulator.jl](https://github.com/alanderos91/BioSimulator.jl)
217//! * [DiffEqJump.jl](https://github.com/SciML/DiffEqJump.jl)
218//! * [Gillespie.jl](https://github.com/sdwfrost/Gillespie.jl)
219//! * [GillespieSSA2](https://github.com/rcannood/GillespieSSA2)
220//! * [Cayenne](https://github.com/quantumbrake/cayenne)
221//!
222//! ## Seem unmaintained
223//!
224//! * [Dizzy](http://magnet.systemsbiology.net/software/Dizzy/)
225//! * [Cellware](http://www.bii.a-star.edu.sg/achievements/applications/cellware/)
226//! * [STOCKS](https://doi.org/10.1093/bioinformatics/18.3.470)
227//! * [StochSim](http://lenoverelab.org/perso/lenov/stochsim.html)
228//! * [Systems biology toolbox](http://www.sbtoolbox.org/)
229//! * [StochKit](https://github.com/StochSS/StochKit) (successor: GillesPy2)
230//! * [SmartCell](http://software.crg.es/smartcell/)
231//! * [NFsim](http://michaelsneddon.net/nfsim/)
232
233pub use rand;
234pub use rand_distr;
235
236mod expr;
237pub mod gillespie;
238mod gillespie_macro;
239
240#[cfg(feature = "pyo3")]
241mod pyo3_gillespie;