# Peroxide
[](https://crates.io/crates/peroxide)
[](http://peroxide.info)
[](https://builds.sr.ht/~axect/Peroxide/.build.yml?)
[](https://travis-ci.org/Axect/Peroxide)


Rust numeric library contains linear algebra, numerical analysis, statistics and machine learning tools with R, MATLAB, Python like macros.
## Why Peroxide?
### 1. Customize features
Peroxide provides various features.
* `default` - Pure Rust (No dependencies of architecture - Perfect cross compilation)
* `O3` - SIMD + OpenBLAS (Perfect performance but hard to set-up - Strongly recommend to read [OpenBLAS for Rust](https://github.com/Axect/Issues/tree/master/Rust))
* `plot` - With matplotlib of python, we can draw any plots.
* `dataframe` - Dataframe & netcdf
* `serde` - serialization with [Serde](https://serde.rs/).
If you want to do high performance computation, then choose openblas feature.
If you don't want to depend C/C++ or Fortran libraries, then choose default feature.
If you want to draw plot with some great templates, then choose plot feature.
You can choose any features simultaneously.
### 2. Easy to optimize
Peroxide uses 1D data structure to describe matrix. So, it's too easy to integrate BLAS & SIMD.
It means peroxide guarantees perfect performance for linear algebraic computations.
### 3. Friendly syntax
Rust is so strange for Numpy, MATLAB, R users. Thus, it's harder to learn the more rusty libraries.
With peroxide, you can do heavy computations with R, Numpy, MATLAB like syntax.
For example,
```rust
extern crate peroxide;
use peroxide::*;
fn main() {
// MATLAB like matrix constructor
let a = ml_matrix("1 2;3 4");
// R like matrix constructor (default)
let b = matrix(c!(1,2,3,4), 2, 2, Row);
// Or use zeros
let mut z = zeros(2, 2);
z[(0,0)] = 1.0;
z[(0,1)] = 2.0;
z[(1,0)] = 3.0;
z[(1,1)] = 4.0;
// Simple but effective operations
let c = a * b; // Matrix multiplication (BLAS integrated)
// Easy to pretty print
c.print();
// c[0] c[1]
// r[0] 1 3
// r[1] 2 4
// Easy to do linear algebra
c.det().print();
c.inv().unwrap().print();
// and etc.
}
```
### 4. Batteries included
Peroxide can do many things.
* Linear Algebra
* Effective Matrix structure
* Transpose, Determinant, Diagonal
* LU Decomposition, Inverse matrix, Block partitioning
* Column, Row operations
* Functional Programming
* More easy functional programming with `Vec<f64>`
* For matrix, there are three maps
* `fmap` : map for all elements
* `col_map` : map for column vectors
* `row_map` : map for row vectors
* Automatic Differentiation
* Dual number system - for 1st order AD
* Hyper dual number system - for 2nd order AD
* Exact jacobian
* `Real` trait to constrain for `f64` and `Dual`
* `Number` structure to unify `f64` and `Dual`
* Numerical Analysis
* Lagrange interpolation
* Cubic spline
* Non-linear regression
* Gradient Descent
* Gauss Newton
* Levenberg Marquardt
* Ordinary Differential Equation
* Euler
* Runge Kutta 4th order
* Backward Euler
* Gauss Legendre 4th order
* Numerical Integration
* Gauss-Legendre Quadrature
* Statistics
* More easy random with `rand` crate
* Ordered Statistics
* Median
* Quantile (Matched with R quantile)
* Probability Distributions
* Bernoulli
* Uniform
* Normal
* Gamma
* Beta
* Student's-t
* RNG algorithms
* Acceptance Rejection
* Marsaglia Polar
* Ziggurat
* Wrapper for `rand-dist` crate
* Special functions
* Wrapper for `special` crate
* Utils
* R-like macro & functions
* Matlab-like macro & functions
* Numpy-like macro & functions
* Julia-like macro & functions
* Plotting
* With `pyo3` & `matplotlib`
* DataFrame
* Convert with Matrix
* Read & Write `csv` files
* Read & Write `netcdf` files
### 5. Written in Rust
Rust & Cargo are awesome for scientific computations.
You can use any external packages easily with Cargo, not make.
And default runtime performance of Rust is also great. If you use many iterations for computations,
then Rust become great choice.
## Latest README version
Corresponding to `0.19.4`
## Pre-requisite
* For `O3` feature - Need `OpenBLAS`
* For `plot` feature - Need `matplotlib` of python
* For `dataframe` feature - Need `netcdf`
## Install
* Add next block to your `cargo.toml`
1. Default
```toml
[dependencies]
peroxide = "0.19"
```
2. OpenBLAS + SIMD
```toml
[dependencies.peroxide]
version = "0.19"
default-features = false
features = ["O3"]
```
3. Plot
```toml
[dependencies.peroxide]
version = "0.19"
default-features = false
features = ["plot"]
```
4. DataFrame
```toml
[dependencies.peroxide]
version = "0.19"
default-features = false
features = ["dataframe"]
```
4. OpenBLAS + SIMD & Plot & DataFrame
```toml
[dependencies.peroxide]
version = "0.19"
default-features = false
features = ["O3", "plot", "dataframe"]
```
## Module Structure
- __src__
- [lib.rs](src/lib.rs) : `mod` and `re-export`
- __macros__ : Macro files
- [julia_macro.rs](src/macros/julia_macro.rs) : Julia like macro
- [matlab_macro.rs](src/macros/matlab_macro.rs) : MATLAB like macro
- [mod.rs](src/macros/mod.rs)
- [r_macro.rs](src/macros/r_macro.rs) : R like macro
- __ml__ : For machine learning (*Beta*)
- [mod.rs](src/ml/mod.rs)
- [reg.rs](src/ml/reg.rs) : Regression tools
- __numerical__ : To do numerical things
- [bdf.rs](src/numerical/bdf.rs) : Backward Differentiation Formula (deprecated)
- [interp.rs](src/numerical/interp.rs) : Interpolation
- [mod.rs](src/numerical/mod.rs)
- [newton.rs](src/numerical/newton.rs) : Newton's Method
- [ode.rs](src/numerical/ode.rs) : Main ODE solver with various algorithms
- [optimize.rs](src/numerical/optimize.rs) : Non-linear regression
- [spline.rs](src/numerical/spline.rs) : Natural Spline
- [utils.rs](src/numerical/utils.rs) : Utils to do numerical things (e.g. jacobian)
- __operation__ : To define general operations
- [extra_ops.rs](src/operation/extra_ops.rs) : Missing operations & Real Trait
- [mut_ops.rs](src/operation/mut_ops.rs) : Mutable operations
- [mod.rs](src/operation/mod.rs)
- [number.rs](src/operation/number.rs) : Number type (include `f64`, `Dual`)
- __redox__ : To wrap `Vec` (Not yet integrated)
- [mod.rs](src/redox/mod.rs)
- [redoxable.rs](src/redox/redoxable.rs)
- __special__ : Wrapper for `special` crate
- [mod.rs](src/special/mod.rs)
- [function.rs](src/special/function.rs) : Special functions
- __statistics__ : Statistical Tools
- [mod.rs](src/statistics/mod.rs)
- [dist.rs](src/statistics/dist.rs) : Probability distributions
- [ops.rs](src/statistics/ops.rs) : Some probabilistic operations
- [rand.rs](src/statistics/rand.rs) : Wrapper for `rand` crate
- [stat.rs](src/statistics/stat.rs) : Statistical tools
- __structure__ : Fundamental data structures
- [dataframe.rs](src/structure/dataframe.rs) : Dataframe
- [dual.rs](src/structure/dual.rs) : Dual number system for automatic differentiation
- [hyper_dual.rs](src/structure/hyper_dual.rs) : Hyper dual number system for automatic differentiation
- [matrix.rs](src/structure/matrix.rs) : Matrix
- [multinomial.rs](src/structure/multinomial.rs) : For multinomial (*Beta*)
- [mod.rs](src/structure/mod.rs)
- [polynomial.rs](src/structure/polynomial.rs) : Polynomial
- [vector.rs](src/structure/vector.rs) : Extra tools for `Vec<f64>`
- __util__
- [mod.rs](src/util/mod.rs)
- [api.rs](src/util/api.rs) : Matrix constructor for various language style
- [non_macro.rs](src/util/non_macro.rs) : Primordial version of macros
- [plot.rs](src/util/plot.rs) : To draw plot (using `pyo3`)
- [print.rs](src/util/print.rs) : To print conveniently
- [useful.rs](src/util/useful.rs) : Useful utils to implement library
- [writer.rs](src/util/writer.rs) : More convenient write system
## Documentation
* [](http://peroxide.info)
## Example
### Basic Runge-Kutta 4th order with inline-python
```rust
#![feature(proc_macro_hygiene)]
extern crate peroxide;
extern crate inline_python;
use peroxide::*;
use inline_python::python;
fn main() {
// Initial condition
let init_state = State::<f64>::new(0f64, c!(1), c!(0));
let mut ode_solver = ExplicitODE::new(test_fn);
ode_solver
.set_method(ExMethod::RK4)
.set_initial_condition(init_state)
.set_step_size(0.01)
.set_times(1000);
let result = ode_solver.integrate();
let x = result.col(0);
let y = result.col(1);
// Plot (Thanks to inline-python)
python! {
import pylab as plt
plt.plot('x, 'y)
plt.show()
}
}
// dy/dx = (5x^2 - y) / e^(x+y)
fn test_fn(st: &mut State<f64>) {
let x = st.param;
let y = &st.value;
let dy = &mut st.deriv;
dy[0] = (5f64 * x.powi(2) - y[0]) / (x + y[0]).exp();
}
```
### Basic Runge-Kutta 4th order with advanced plotting
```rust
extern crate peroxide;
use peroxide::*;
fn main() {
let init_state = State::<f64>::new(0f64, c!(1), c!(0));
let mut ode_solver = ExplicitODE::new(test_fn);
ode_solver
.set_method(ExMethod::RK4)
.set_initial_condition(init_state)
.set_step_size(0.01)
.set_times(1000);
let result = ode_solver.integrate();
let x = result.col(0);
let y = result.col(1);
// Plot (using python matplotlib)
let mut plt = Plot2D::new();
plt.set_domain(x)
.insert_image(y)
.set_title("Test Figure")
.set_fig_size((10, 6))
.set_dpi(300)
.set_legend(vec!["RK4"])
.set_path("example_data/test_plot.png");
// Remove below comments to activate
//plt.savefig();
}
fn test_fn(st: &mut State<f64>) {
let x = st.param;
let y = &st.value;
let dy = &mut st.deriv;
dy[0] = (5f64 * x.powi(2) - y[0]) / (x + y[0]).exp();
}
```
### Basic Runge-Kutta 4th order with exporting netcdf
```rust
extern crate peroxide;
use peroxide::*;
fn main() {
let init_state = State::<f64>::new(0f64, c!(1), c!(0));
let mut ode_solver = ExplicitODE::new(test_fn);
ode_solver
.set_method(ExMethod::RK4)
.set_initial_condition(init_state)
.set_step_size(0.01)
.set_times(1000);
let result = ode_solver.integrate();
let x = result.col(0);
let y = result.col(1);
// Construct DataFrame
let mut df = DataFrame::with_headers(vec!["x", "y"]);
df["x"] = x;
df["y"] = y;
// Write netcdf (Remove below comment)
//df.write_nc("example_data/rk4_test.nc").expect("Can't write nc files");
}
fn test_fn(st: &mut State<f64>) {
let x = st.param;
let y = &st.value;
let dy = &mut st.deriv;
dy[0] = (5f64 * x.powi(2) - y[0]) / (x + y[0]).exp();
}
```
### Basic Runge-Kutta 4th order with Stop condition
```rust
extern crate peroxide;
use peroxide::*;
fn main() {
let init_state = State::<f64>::new(0f64, c!(1), c!(0));
let mut ode_solver = ExplicitODE::new(test_fn);
ode_solver
.set_method(ExMethod::RK4)
.set_initial_condition(init_state)
.set_step_size(0.01)
.set_stop_condition(stop) // Add stop condition
.set_times(1000);
let result = ode_solver.integrate();
let x = result.col(0);
let y = result.col(1);
let mut plt = Plot2D::new();
plt.set_domain(x)
.insert_image(y)
.set_title("Test Figure")
.set_fig_size((10, 6))
.set_dpi(300)
.set_legend(vec!["RK4"])
.set_path("example_data/test_plot.png");
plt.savefig();
}
fn test_fn(st: &mut State<f64>) {
let x = st.param;
let y = &st.value;
let dy = &mut st.deriv;
dy[0] = (5f64 * x.powi(2) - y[0]) / (x + y[0]).exp();
}
fn stop(st: &ExplicitODE) -> bool {
let y = &st.get_state().value[0];
(*y - 2.4).abs() < 0.01
}
```

### Multi-Layer Perceptron (from scratch)
```rust
extern crate peroxide;
use peroxide::*;
// x : n x L
// xb: n x (L+1)
// v : (L+1) x M
// a : n x M
// ab: n x (M+1)
// w : (M+1) x n
// wb: M x N
// y : n x N
// t : n x N
// dh: n x M
// do: n x N
fn main() {
let v = weights_init(3, 2);
let w = weights_init(3, 1);
let x = ml_matrix("0 0; 0 1; 1 0; 1 1");
let t = ml_matrix("0;1;1;0");
let y = train(v, w, x, t, 0.25, 5000);
y.print();
}
fn weights_init(m: usize, n: usize) -> Matrix {
rand(m, n) * 2f64 - 1f64
}
fn sigmoid(x: f64) -> f64 {
1f64 / (1f64 + (-x).exp())
}
fn forward(weights: Matrix, input_bias: Matrix) -> Matrix {
let s = input_bias * weights;
s.fmap(|x| sigmoid(x))
}
fn add_bias(input: Matrix, bias: f64) -> Matrix {
let b = matrix(vec![bias; input.row], input.row, 1, Col);
cbind(b, input)
}
fn hide_bias(weight: Matrix) -> Matrix {
weight.skip(1, Row)
}
fn train(
weights1: Matrix,
weights2: Matrix,
input: Matrix,
answer: Matrix,
eta: f64,
times: usize,
) -> Matrix {
let x = input;
let mut v = weights1;
let mut w = weights2;
let t = answer;
let xb = add_bias(x.clone(), -1f64);
for _i in 0..times {
let a = forward(v.clone(), xb.clone());
let ab = add_bias(a.clone(), -1f64);
let y = forward(w.clone(), ab.clone());
// let err = (y.clone() - t.clone()).t() * (y.clone() - t.clone());
let wb = hide_bias(w.clone());
let delta_o = (y.clone() - t.clone()) * y.clone() * (1f64 - y.clone());
let delta_h = (delta_o.clone() * wb.t()) * a.clone() * (1f64 - a.clone());
w = w.clone() - eta * (ab.t() * delta_o);
v = v.clone() - eta * (xb.t() * delta_h);
}
let a = forward(v, xb);
let ab = add_bias(a, -1f64);
let y = forward(w, ab);
y
}
```
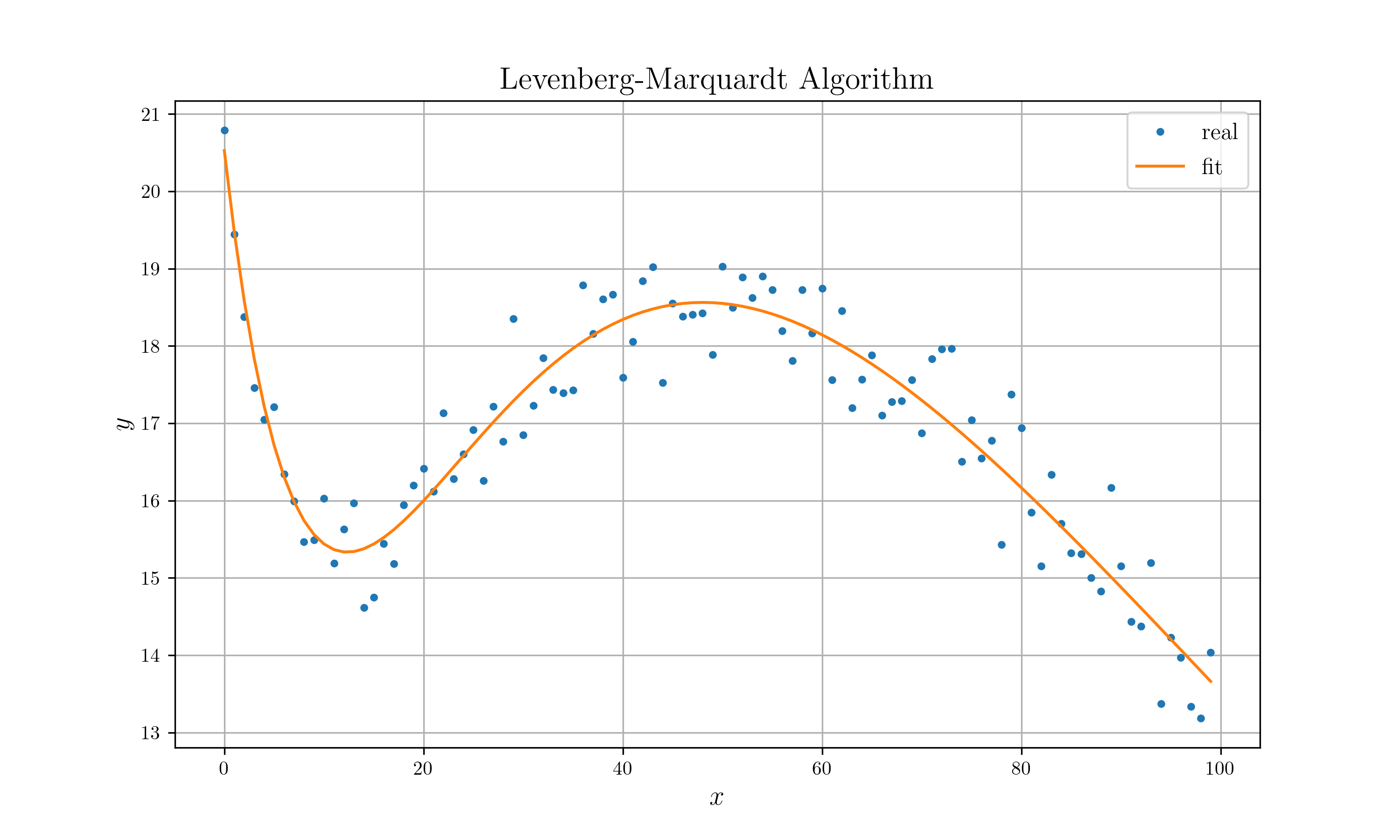
### Levenberg-Marquardt Algorithm (refer to [lm.pdf](http://people.duke.edu/~hpgavin/ce281/lm.pdf))
```rust
extern crate peroxide;
use peroxide::*;
fn main() {
let noise = Normal(0., 0.5);
let p_true: Vec<Number> = NumberVector::from_f64_vec(vec![20f64, 10f64, 1f64, 50f64]);
let p_init = vec![5f64, 2f64, 0.2f64, 10f64];
let domain = seq(0, 99, 1);
let real = f(&domain, p_true.clone()).to_f64_vec();
let y = zip_with(|x, y| x + y, &real, &noise.sample(100));
let data = hstack!(domain.clone(), y.clone());
let mut opt = Optimizer::new(data, f);
let p = opt
.set_init_param(p_init)
.set_max_iter(100)
.set_method(LevenbergMarquardt)
.optimize();
p.print();
opt.get_error().print();
let mut plt = Plot2D::new();
plt.set_domain(domain)
.insert_image(y)
.insert_image(p)
.set_legend(vec!["real", "fit"])
.set_title("Levenberg-Marquardt Algorithm")
.set_path("example_data/lm_test.png")
.set_marker(vec![Point, Line])
.savefig()
.expect("Can't draw a plot");
}
fn f(domain: &Vec<f64>, p: Vec<Number>) -> Option<Vec<Number>> {
Some(
domain.clone().into_iter()
.map(|t| Number::from_f64(t))
.map(|t| p[0] * (-t / p[1]).exp() + p[2] * t * (-t / p[3]).exp())
.collect()
)
}
```
## Version Info
To see [RELEASES.md](./RELEASES.md)
## Contributes Guide
See [CONTRIBUTES.md](./CONTRIBUTES.md)
## TODO
To see [TODO.md](./TODO.md)