# INTRODUCING NUCLEOB
[nucleob](https://github.com/piotrbajdek/nucleob) is a tool that performs statistical analysis on .fasta files containing nucleotide or amino acid sequences. It is engineered to produce highly reliable, clearly formatted results while maintaining a minimalistic design and cross-platform compatibility.
[keywords: amino acids, bioinformatics, biology, DNA, fasta, genes, genetics, molecular, nucleic acids, nucleobases, nucleotides, proteins, RNA, scientific computing, statistics, stats]
# EXAMPLES
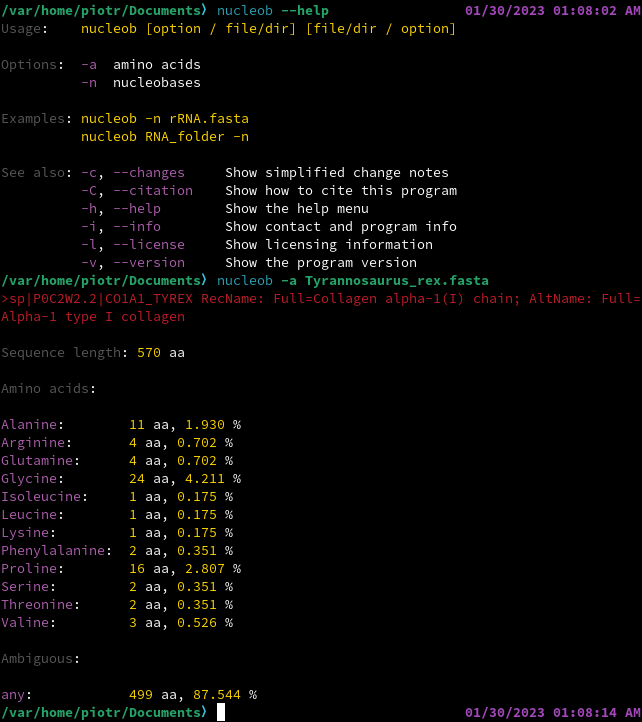
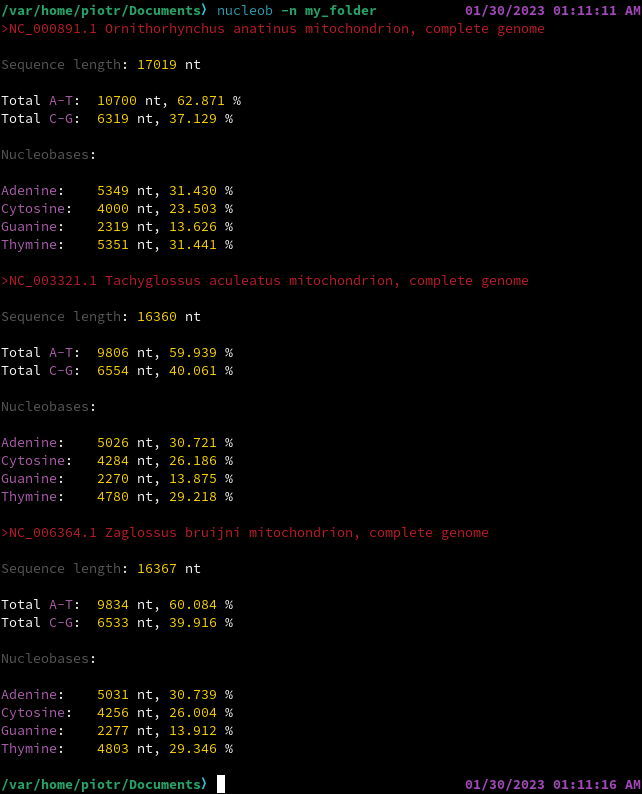
# CITATION AND REUSE
nucleob can be modified and forked under the terms of the [MIT License](https://github.com/piotrbajdek/nucleob/blob/main/LICENSE.md). When citing nucleob in research articles, it is important to refer to a specific program version, such as:
Bajdek, P., 2023. nucleob (version 1.1.1). [computer software] https://github.com/piotrbajdek/nucleob
# INSTALLATION ON LINUX
[nucleob](https://github.com/piotrbajdek/nucleob) is designed to be compatible with **Windows** and **macOS**, and can be easily installed using [cargo](https://www.rust-lang.org/tools/install). However, the primary development and testing environment for nucleob is **Fedora Linux**.
The current version of nucleob (v1.1.1) has been verified to work properly on Fedora Linux 37 and Ubuntu 22.10.
## METHOD 1 – USING CARGO
**[Recommended for programmers]**
**1.** To install nucleob from [crates.io](https://crates.io/crates/nucleob), use the following cargo command:
_cargo install nucleob_
The executable will be saved in the hidden `.cargo/bin/` directory within your home directory.
**2a.** For easy access, you may want to copy the nucleob file to the `/usr/bin/` directory. This can be done by following the instructions in Method 2 (3a, 3b).
**2b.** As an alternative, you can add the `~/.cargo/bin/` directory to your system's PATH variable, which can be configured using [rustup](https://www.rust-lang.org/tools/install).
## METHOD 2 – UNIVERSAL LINUX BINARIES
**1.** To install nucleob, first download the distro-independent [binary](https://github.com/piotrbajdek/nucleob/releases/download/v1.1.1/nucleob) from GitHub.
**2.** Then, make the file executable by running the command:
_sudo chmod +x ./nucleob_
**3a.** On most Linux distributions, install nucleob by copying the binary to `/usr/bin/`:
_sudo cp nucleob /usr/bin/_
**3b.** For Fedora Silverblue / Kinoite, use this command:
_sudo cp nucleob /var/usrlocal/bin/_
## METHOD 3 – DISTRO-SPECIFIC PACKAGES
**[Recommended for most users]**
Distro-specific packages for [.rpm](https://github.com/piotrbajdek/nucleob/releases/download/v1.1.1/nucleob-1.1.1-1.x86_64.rpm) and [.deb](https://github.com/piotrbajdek/nucleob/releases/download/v1.1.1/nucleob_1.1.1_amd64.deb)-based Linux distributions are also available for download. To install nucleob on different Linux distributions, follow these instructions:
Fedora Linux / RHEL / openSUSE:
_sudo rpm -i nucleob-1.1.1-1.x86_64.rpm_
Fedora Silverblue / Kinoite:
_rpm-ostree install nucleob-1.1.1-1.x86_64.rpm_
Ubuntu:
_sudo dpkg -i nucleob_1.1.1_amd64.deb_
## METHOD 4 – MANUAL COMPILATION
First, download and unpack the nucleob [source code](https://github.com/piotrbajdek/nucleob/archive/refs/tags/v1.1.1.zip) from GitHub. Next, to build and install the program, use the command:
_cargo build \--release && sudo cp target/release/nucleob /usr/bin/_