Expand description
To optimize parametric model (non-linear regression)
§Optimizer structure
§Declaration
extern crate peroxide;
use peroxide::fuga::*;
use std::collections::HashMap;
pub struct Optimizer<F>
where F: Fn(&Vec<f64>, Vec<AD>) -> Option<Vec<AD>> {
domain: Vec<f64>,
observed: Vec<f64>,
func: Box<F>,
param: Vec<AD>,
max_iter: usize,
error: f64,
method: OptMethod,
option: HashMap<OptOption, bool>,
hyperparams: HashMap<String, f64>,
}§Method (Should fill)
new: Declare new Optimizer. Should be mutableset_init_param: Input initial parameterset_max_iter: Set maximum number of iterationsset_method: Set method to optimization
§Method (Optional)
get_domain: Get domainget_error: Root mean square errorget_hyperparam: Get hyperparameterset_lr: Set learning rate (ForGradientDescent)set_lambda_init: Set initial value of lambda (ForLevenbergMarquardt)set_lambda_max: Set maximum value of lambda (ForLevenbergMarquardt)
§Method (Generate result)
optimize: Optimize
§Example
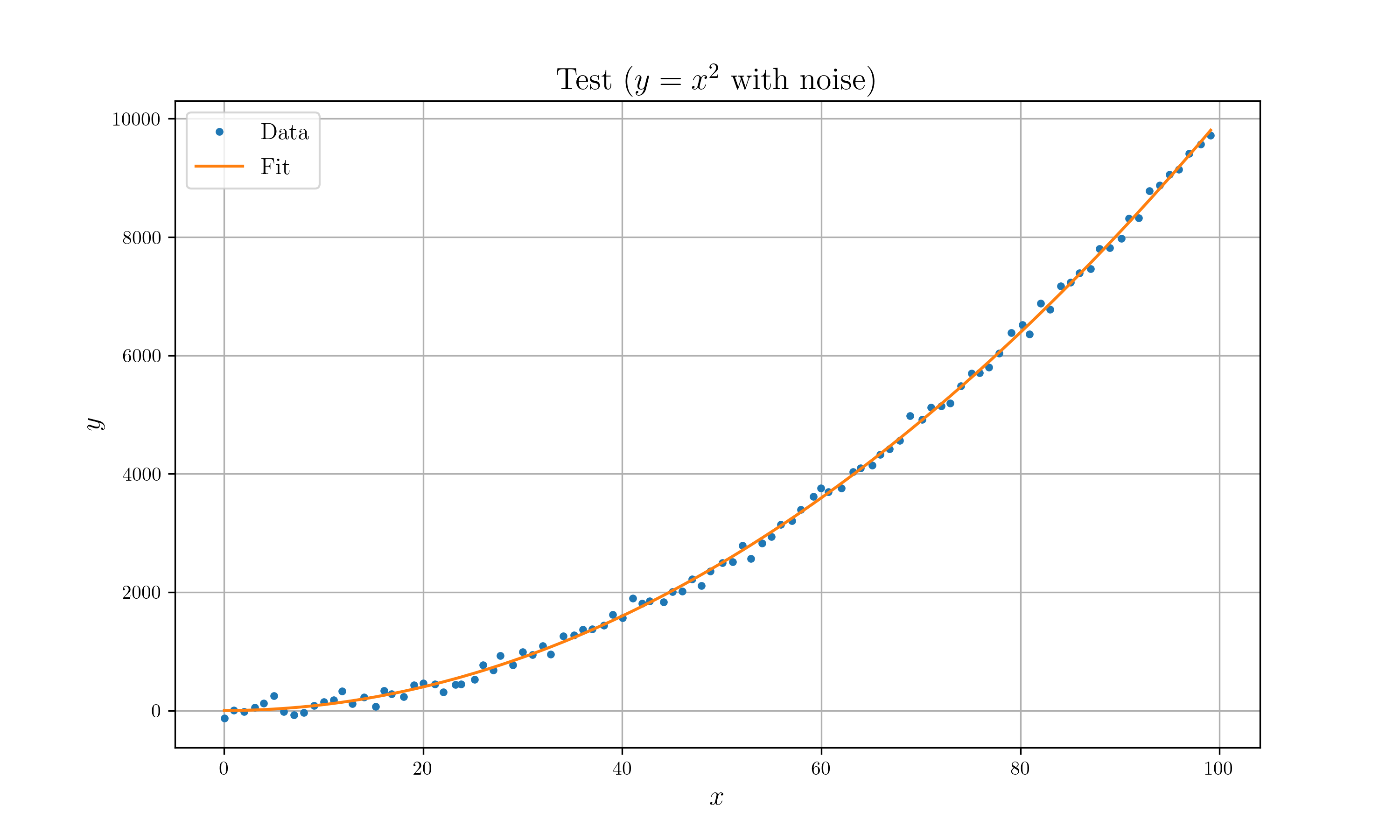
- Optimize $y = x^n$ model with $y = x^2$ with gaussian noise.
#[macro_use]
extern crate peroxide;
use peroxide::fuga::*;
fn main() {
// To prepare noise
let normal = Normal(0f64, 0.1f64);
let normal2 = Normal(0f64, 100f64);
// Noise to domain
let mut x = seq(0., 99., 1f64);
x = zip_with(|a, b| (a + b).abs(), &x, &normal.sample(x.len()));
// Noise to image
let mut y = x.fmap(|t| t.powi(2));
y = zip_with(|a, b| a + b, &y, &normal2.sample(y.len()));
// Initial parameter
let n_init = vec![1f64];
let data = hstack!(x.clone(), y.clone());
// Optimizer setting
let mut opt = Optimizer::new(data, quad);
let p = opt.set_init_param(n_init)
.set_max_iter(50)
.set_method(LevenbergMarquardt)
.set_lambda_init(1e-3) // Optional: Set initial value of lambda (Only for `LevenbergMarquardt`)
.set_lambda_max(1e+3) // Optional: Set maximum bound of lambda (Only for `LevenbergMarquardt`)
.optimize();
p.print(); // Optimized parameter
opt.get_error().print(); // Optimized RMSE
// Plot
//#[cfg(feature = "plot")]
//{
// let z = quad(&x, p.to_ad_vec()).unwrap().to_f64_vec();
// let mut plt = Plot2D::new();
// plt.set_domain(x)
// .insert_image(y) // plot data
// .insert_image(z) // plot fit
// .set_legend(vec!["Data", "Fit"])
// .set_title("Test ($y=x^2$ with noise)")
// .set_path("example_data/lm_test.png")
// .set_marker(vec![Point, Line])
// .savefig().expect("Can't draw a plot");
//}
}
fn quad(x: &Vec<f64>, n: Vec<AD>) -> Option<Vec<AD>> {
Some(
x.clone().into_iter()
.map(|t| AD1(t, 0f64))
.map(|t| t.pow(n[0]))
.collect()
)
}
Re-exports§
pub use self::OptMethod::GaussNewton;pub use self::OptMethod::GradientDescent;pub use self::OptMethod::LevenbergMarquardt;
Structs§
- Optimizer for optimization (non-linear regression)